ABSTRACT
Recent developments in precision oncology have increased the complexity of diagnostic and therapeutic decisions. Here, we broadly review the field of precision oncology and discuss common mutational drivers in non–small cell lung cancer (NSCLC) that directly relate to the diagnosis, evaluation, and treatment of patients with metastatic disease.
A number of driver alterations (mutations and chromosomal rearrangements) occur in patients with NSCLC.
Mutations in the EGFR and BRAF genes and rearrangements involving the ALK and ROS1 genes can be targeted with novel agents.
These targeted therapies have demonstrated superior outcomes and far less toxicity compared with traditional cytotoxic chemotherapy in patients with metastatic NSCLC.
Efficiently identifying genetic alterations that can be treated with existing therapies is key to providing best-practice care to all patients.
In the past few years, targeted therapies have become widely available and have revolutionized the treatment of patients with advanced solid tumors, particularly metastatic non–small cell lung cancer (NSCLC). For patients who have 1 of a select few actionable genetic alterations, phase 3 trials in NSCLC have consistently shown survival benefits associated with targeted agents compared with chemotherapy.1–3 Large-scale real-world data suggest these targeted therapies are improving survival on a population level.4
Targeted therapies are costly, with estimates of cost per quality-adjusted life-year of $150,000 to over $200,000. However, they are also associated with improved quality of life and fewer adverse effects compared with chemotherapy.3,5–8
The drugs fall under the expanding umbrella term of “precision oncology,” which refers to both the diagnostic method (ie, genomic sequencing) and the treatments prescribed based on the results. Recent advances in genomic sequencing have allowed for efficient and reliable identification of patients who may benefit from precision therapies.
Here, we review precision oncology and the most clinically relevant mutations that can be found among patients with metastatic NSCLC. We further review the diagnostic tests available to clinicians to assess for these mutations. Last, we discuss opportunities to streamline testing in an efficient manner.
PRECISION ONCOLOGY
Advances in the diagnosis and treatment of NSCLC have come to define the paradigm of precision oncology (Figure 1). Through remarkable laboratory-based efforts and wide-ranging epidemiologic studies, a significant number of critical genetic alterations that cause cells to grow, divide, and turn cancerous have been discovered.
The current paradigm for precision oncology for NSCLC.
As opposed to other accompanying and functionally neutral (“passenger”) mutations, these specific “driver” mutations are functionally important to the growth of the malignancy.9 Further investigation into these driver mutations uncovered targeted therapies that provide a line of highly efficacious treatments, significantly improving overall survival for patients with metastatic NSCLC.
These developments have fundamentally altered clinicians’ approaches to intervention in NSCLC over the past decade. Additionally, successes achieved in patients with NSCLC have encouraged further research efforts toward expanding the role of precision oncology for patients with other advanced malignancies.
In this review, we do not discuss immunotherapy, which is a general term referring to immune checkpoint inhibitors, namely agents that alter the cytotoxic T-lymphocyte– associated protein 4 and programmed death-ligand 1 pathways. These agents have also vastly reshaped the treatment paradigm for patients with metastatic NSCLC, but specifically have a far greater role in patients who do not have a highly actionable mutation or fusion. The topic of immunotherapy is part of a broader discussion than is possible in this review.
GENETIC ALTERATIONS FOR WHICH THERAPIES ARE APPROVED
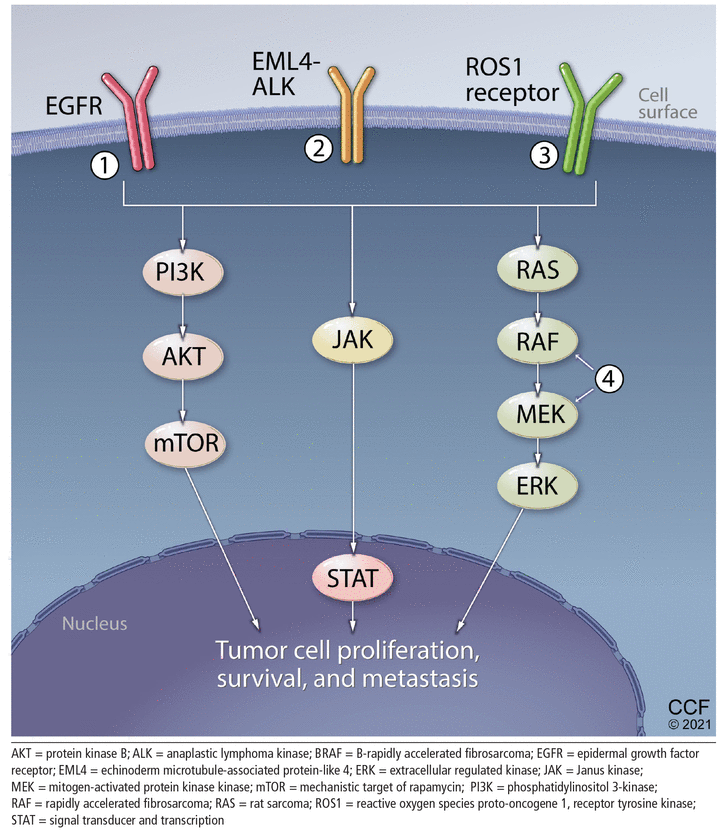
Several genetic alterations identified in patients with metastatic NSCLC can currently be targeted with therapies approved by the US Food and Drug Administration (FDA), including mutations in the epidermal growth factor receptor (EGFR) and BRAF genes and chromosomal rearrangements of the anaplastic lymphoma kinase (ALK) and ROS1 genes. The rates of alterations are shown in Figures 1 and 2. The associated targeted therapies for the different alterations are described in Table 110–22 and Figure 2. Definitions and examples of key terms used in this article are given in Table 2, while a schematic review of the consequences of various actionable alterations is shown in Figure 3.
Rates of actionable mutations in patients with non–small cell lung cancer (NSCLC). Of note, NSCLC encompasses about 85% of lung cancers. Compared with smokers, nonsmokers have far higher rates of actionable mutations.
aThough another 20% to 30% of patients with NSCLC have some form of actionable alteration, the corresponding targeted agents are not necessarily FDA-approved. Of note, drugs targeting MET and RET have recently been approved for suitable NSCLC candidates.
Approved targeted therapies for non–small cell lung cancer and their comparative effectiveness
Definitions and descriptions of key terms
Pathways of proliferation. Certain key proteins that are abnormally active due to mutations and genetic rearrangements contribute to tumor cell proliferation, survival, and metastasis. Targeted therapies can block these pathways, specifically inhibitors of (1) epidermal growth factor receptor (EGFR), (2) anaplastic lymphoma kinase (ALK), (3) ROS1, (4) BRAF/MEK, and others.
EGFR mutations
EGFR is a transmembrane tyrosine kinase receptor that operates within signal transduction pathways facilitating cellular growth and apoptosis. In the United States, nearly 20% of patients with NSCLC harbor a pathogenic EGFR mutation.23 Mutations in the EGFR gene, which codes for the EGFR receptor, lead to dimerization of receptors. This dimerization causes constitutive activity of the tyrosine kinase associated with the EGFR protein, thereby inducing a hyperproliferative state.
Targeted treatments are directed toward inhibiting either the extracellular receptor or the intracellular tyrosine kinase. Among patients with metastatic NSCLC, efforts to inhibit intracellular tyrosine kinase have been most successful. The following drugs that inhibit EGFR tyrosine kinase are FDA-approved:
Erlotinib, a first-generation drug
Gefitinib, a first-generation drug
Afatinib, a second-generation drug
Dacomitinib, a second-generation drug
Osimertinib, a third-generation drug.
A number of mutations can be found within the EGFR gene. The variants that are most susceptible to targeted treatments include exon 19 deletions and exon 21 substitutions (L858R). Cancers associated with less common mutations involving exon 18 and 20 may respond to tyrosine kinase inhibitor (TKI)-based therapy, but sensitivity varies by specific mutation and is often lower compared with exon 19 and 21 mutations.
A number of clinical trials have demonstrated marked improvements in overall survival with use of TKIs compared with traditional chemotherapy in patients with an EGFR mutation. Later-generation TKIs such as osimertinib not only overcome a common mechanism of resistance, the T790M mutation, but also provide better progression-free and overall survival outcomes than earlier-generation TKIs for all patients with metastatic NSCLC harboring typical pathogenic EGFR mutations.24
Common adverse effects with TKIs are predominantly cutaneous, namely acneiform rash and dry skin, followed by diarrhea. Rarely, patients may develop interstitial lung disease. This is not an exhaustive list of potential adverse effects and neither are the adverse effect profiles described for the targeted therapies listed for patients harboring actionable alterations in ALK, ROS1, or BRAF.
BRAF mutations
BRAF mutations, commonly associated with melanoma, lead to a mutated serine-threonine kinase in the MAPK kinase pathway. A BRAF mutation is the driver oncogene in 1% to 3% of cases of NSCLC.25
NSCLC BRAF mutations take multiple forms, including the classic V600E form (50%), a G469A form (40%), and a D594G form (11%). Targeted therapies developed to date are primarily effective against the V600E mutation. Specific targeting of MEK1/2 mutations further downstream in the signaling pathway has also demonstrated long-term benefit and has been approved as a treatment option by the FDA.
Currently available and approved therapies for BRAF-mutant NSCLC include:
Dabrafenib, a V600E serine/threonine kinase inhibitor
Trametinib, a MEK 1/2 inhibitor, used in combination with dabrafenib.
Additional therapies being investigated include a combination of encorafenib with binimetinib, among others.
Common side effects of BRAF and MEK inhibitors include rash, diarrhea, and fever. A wide collection of uncommon adverse effects have been described, including systolic heart failure and retinopathy.
ALK rearrangements
ALK rearrangements lead to fusion protein products, most commonly involving echinoderm microtubule protein-like 4 (EML4). In the United States, nearly 6% of patients with NSCLC harbor an ALK rearrangement.23 The fusion in these rearrangements connects the ALK protein with exon 20 of the EML4 protein, thereby leading to constitutive activation of the ALK tyrosine kinase. Similar to EGFR mutations, the ALK rearrangement creates a downstream transduction pathway via the AKT and ERK signaling pathways that encourages growth and discourages apoptosis.
ALK inhibitors have demonstrated excellent outcomes among patients with metastatic ALK-rearranged NSCLC.
Common adverse effects with ALK inhibitors include gastrointestinal toxicities. Bradycardia, QT prolongation, and interstitial lung disease are possible.
Currently available and approved ALK inhibitors are:
Crizotinib, a first-generation drug
Ceritinib, a first-generation drug
Alectinib, a second-generation drug
Brigatinib, a second-generation drug
Lorlatinib, a third-generation drug.
ROS1 rearrangements
Rearrangements of the receptor tyrosine kinase c-ros oncogene 1 (ROS1) on chromosome 6 lead to constitutive tyrosine kinase activity, stimulating oncogenic signals through downstream pathways. Importantly, the ROS1 rearrangements result in a mutant protein form that is structurally very similar to that seen among ALK rearrangements. That structural similarity creates cross-sensitivity and cross-reactivity with broad-target tyrosine kinase inhibitors, allowing for use of these targeted therapies in patients with ROS1 rearrangements in addition to their originally intended targets. Approximately 1% of patients with NSCLC in the United States harbor a ROS1 rearrangement.
Currently approved therapies include:
Crizotinib, first-generation
Entrectinib, first-generation.
Other tyrosine kinase inhibitors in development or recommended as alternative therapies include ceritinib.
Adverse effects are drug-dependent. Targeted agents that concurrently serve as ALK inhibitors, such as crizotinib, share the aforementioned ALK-inhibitor risk profiles. On the other hand, entrectinib is part of a separate collection of drugs that are typically prescribed for patients with neurotrophic tyrosine receptor kinase (NTRK) gene fusions in other solid tumors. Patients receiving these drugs may face a separate group of adverse effects, most commonly fatigue, liver and kidney dysfunction, and myelosuppression.
MET, RET, AND OTHERS
In the summer of 2020, the FDA approved treatments for patients harboring alterations in RET (selpercatinib and pralsetinib) and MET (capmatinib).26–28
However, these alterations represent only a fraction of the spectrum of pathogenic alterations in NSCLC; many more are currently being investigated in the laboratory and through clinical research. These include alterations in KRAS, NRAS, AKT, DDR2, HER2 (ERBB2), PIK3CA, MEK1, PTEN, and FGFR.29,30
This list, and our understanding of how these alterations drive tumorigenesis in NSCLC, will continue to expand in the years to come.
TESTS FOR CLINICALLY RELEVANT MUTATIONS
Precision oncology requires equal emphasis on new drugs and identifying the patients most likely to benefit from them. Medical oncologists constantly face decisions about the best diagnostic test and timing of testing for their patients with NSCLC. A thorough understanding of the tests available is therefore critically important to the delivery of the best possible care.
The current diagnostic tests include:
Immunohistochemical (IHC) staining
Fluorescence in situ hybridization (FISH)
Reverse transcriptase polymerase chain reaction (RT-PCR)
Tissue-based next-generation sequencing (NGS).
The diagnostic accuracy, breadth of mutations, financial cost, and time required for each test vary considerably.
Immunohistochemical staining
IHC staining is a histology-based analytical tool for identifying mutational variants through specialized stains and targeted antibodies to demonstrate the presence or absence of a genetic variant within the tissue sample. It is largely used as a screening tool, given its demonstrated ability to efficiently capture identifiable variants.
Multiple studies have demonstrated sensitivity ranging from 86% to 100% and specificity of 76% to 100% for detecting ALK variants, with similar evidence for detecting EGFR mutations.31–35
The cost of IHC ranges from $33 to $124 and averages $73, making it the cheapest test for mutations.36 IHC testing for ALK is FDA-approved, with FISH used for equivocal cases. For ROS1, IHC may be used in screening, but further FISH, PCR, or NGS testing should be used to confirm positive results and rule out false-positive results.
Because of limited sensitivity in detecting specific EGFR mutations, using IHC to determine candidacy for targeted agents against EGFR is discouraged in current guidelines for mutational testing.37
Fluorescence in situ hybridization
Similar to IHC, FISH analysis utilizes patient tissue samples for a histology-based assay of genetic variants. However, unlike IHC, FISH probes are predicated on complementary binding that can identify specific genetic sequences of interest. Using fluorescently labeled DNA or RNA probes created to reciprocally bind targets of interest, FISH analyses can detect the presence of their target sequences, and thus genetic variants, within prepared tissue samples.
FISH remains the gold standard for detecting mutant fusion protein variants and is still widely used for this purpose today. The sensitivity ranges between 90.3% and 100% and the specificity between 97.7% and 100% among patients being tested for ALK rearrangements.38,39 Cost of FISH testing averages about $300, and turnaround processing time averages about 2 to 5 days, marginally longer than that of IHC processing.36
The most significant drawbacks of FISH testing arise from its limited scope (each test is specific for 1 genetic variant), need for fluorescent microscope workstations, and the qualitative component of its assessment (there may be some ambiguity based on the cutoff point for positive vs negative results).40
Reverse transcriptase polymerase chain reaction-based methods
RT-PCR analysis uses unique, labeled DNA probes to identify, amplify, and quantify the levels of specific genetic variants in tissue samples. It has demonstrated efficacy and accuracy as a stand-alone diagnostic tool and in comparison to IHC, FISH, and NGS.41 Advantages: it can perform multiple simultaneous assessments, it can be done on samples other than biopsy tissue (such as blood), and it is objective—there is no subjective rating of positivity as in IHC and FISH. Its sensitivity for identifying mutational variants ranges from 88% to 100% and its specificity from 94% to 100%.39,42,43
While the individual costs of a single RT-PCR assay are difficult to characterize owing to the variability of pricing of reagents, technical labor, and available facilities, multiple studies have demonstrated the cost-effectiveness of RT-PCR testing in comparison to histology-based diagnostic tools.
Tissue-based next-generation sequencing
By identifying the full genetic sequences of targeted areas of the genome, NGS is able to identify both documented and previously undiscovered mutational variants by similar principles of complementary nucleotide binding as RT-PCR, but at a larger scale.44 This broad applicability allows for interrogation of an ever-expanding library of driver mutations, all at once, with pinpoint accuracy.
Advances in NGS technology over the last several years have driven down overall costs while improving accuracy and ease of application, making economical feasibility a reality. NGS is now commonly used in genetic assessment in advanced NSCLC.45–47 In its earliest iterations, NGS was demonstrated to have high sensitivity and specificity values by validation studies (95%–99%, with positive predictive value > 99%).48 More recent studies have assessed these markers of accuracy at 100% for both sensitivity and specificity, establishing NGS as the comparative technique against which other mutation identification processes can be evaluated.49,50
However, the estimated cost of targeted gene panel sequencing averages $1,609, with significant variation depending on the size of the panel of mutational targets, preference for whole-exome sequencing ($4,459), or whole-exome plus RNA sequencing ($5,938).45 In addition, turnaround times for NGS studies are long, with estimates of 13 to 21 days on average in multiple studies.51
Plasma genotyping
Plasma genotyping, popularly called “liquid biopsy,” a broad collection of screening tests utilizing capture and identification of circulating tumor DNA (ctDNA), has demonstrated incredible promise in its early forms.52–55 It has significant clinical potential, given its ease of implementation, low risk compared with tis sue-dependent screening methods, rapid turnaround time, and ability to perform screening analysis without limitations (eg, amount of tissue collected, need for repeat biopsy). This technology may allow for detection of new actionable mutations, characterization of response to therapy, and identification of mechanisms of resistance to therapy.56,57 Early assessments have demonstrated some level of agreement between ctDNA assessments and previously confirmed tissue diagnoses, with high levels of individualized variant identification by ctDNA alone.
HOW HAS NEXT-GENERATION SEQUENCING ALTERED TESTING PRACTICES?
Clinicians practicing precision medicine must carefully consider the cost-benefit analysis of this approach and plan their diagnostic and therapeutic course accordingly: What actionable information will result from testing? What testing method will provide maximal utilizable information at the lowest cost? What is the feasibility of implementing a therapy based on that information?
Clinicians can use a wide array of testing procedures that have well-documented clinical efficacy, from histology-based IHC analyses to small-scale quantitative PCR assays. Employing these tests for initial screening, especially in settings with limited access to advanced technologies or ability to follow through on the data they provide, even for a faster stepwise diagnostic approach, could allow clinical oncologists to refine their approach to diagnosis and treatment in the precision medicine era.
NGS technology provides an unparalleled view of the genetic framework of a patient’s disease. It allows clinicians and researchers to identify a significant proportion of the full mutational burden of a tumor and uncover the various targets for which therapies can be used. This has created many opportunities for research and clinical investigation of this technology, opening the door for trials exploring the efficacy of a wide range of therapies.
Looking ahead, application of NGS technology to ctDNA isolated from simple blood samples continues to expand the landscape of precision medicine. The potential to identify and exhaustively characterize tumors with rapid, noninvasive diagnostic tools is incredibly appealing. Like NGS technology and the precision oncology movement as a whole, the inherent potential for paradigm-shifting clinical impact will continue to drive interest in this technology.
FUTURE DIRECTIONS FOR RESEARCH AND CLINICAL PRACTICE
As research advances our understanding of the molecular framework of NSCLC, clinicians must stay informed about the latest testing methods and therapies, actionable mutations, and breakthrough approaches. Research into the EGFR, BRAF, ALK, ROS1, and other alterations driving disease has unlocked treatments that have changed the course of disease in countless patients. The use of precision medicine in NSCLC will benefit patients for years to come.
Future discussions of the research and therapies surrounding NSCLC will necessarily focus on:
Discovery of new driver mutations
New therapies that target these currently unidentified mutations
Advances in currently developed therapies
Results of clinical trials and bench research currently in progress
Expansion and streamlining of the testing procedures used for variant identification (ie, genomic sequencing).
DISCLOSURES
Dr. Clarke reports financial relationships (consulting, independent contracting, or research) with AbbVie Pharmaceuticals, Adaptimmune, Array Bio-Pharma, AstraZeneca, Bristol Myers Squibb, Genentech, Grid Therapeutics, Guardant Health, Merck, Moderna, NGM Biopharmaceuticals, Pfizer, and Spectrum Pharmaceuticals.
All other authors report no relevant financial relationships which, in the context of their contributions, could be perceived as a potential conflict of interest.
- Copyright © 2021 The Cleveland Clinic Foundation. All Rights Reserved.