This article aims to introduce the statistical methodology behind chi-square and Fisher’s exact tests, which are commonly used in medical research to assess associations between categorical variables. This discussion will use data from a study by Mrozek1 in patients with acute respiratory distress syndrome (ARDS). This was a multicenter, prospective, observational study: multicenter because it included data from 10 intensive care units, prospective because the study collected the data moving forward in time, and observational because the study investigators did not have control over the group assignments but rather used the naturally occurring groups. The study objective was to characterize focal and nonfocal patterns of lung computed tomography (CT)-based imaging with plasma markers of lung injury.
The primary grouping variable was type of ARDS (focal vs nonfocal) as determined by CT scans and other lung imaging tools. In this study, there were 32 (27%) patients with focal ARDS and 87 (73%) patients with nonfocal ARDS. What will be important, however, is classifying the type of variables because this determines the type of analyses performed. Type of ARDS is a categorical variable with 2 levels.
The primary study endpoint was plasma levels of the soluble form of the receptor for advanced glycation end product. There were also a number of secondary study endpoints that can be grouped as either patient outcomes or biomarkers. Patient outcomes included the duration of mechanical ventilation and both 28- and 90-day mortality. Levels of other biomarkers included surfactant protein D, soluble intercellular adhesion molecule-1, and plasminogen activator inhibitor-1.
This article focused on the secondary outcome of 90-day mortality beginning at disease onset. Again, we are interested in classifying this variable, which is categorical with 2 levels (yes vs no). So the scenario is that we want to assess the relationship between the type of ARDS (focal vs nonfocal) and 90-day mortality (yes vs no). In its most basic form, this scenario is an investigation into the association among 2 categorical variables.
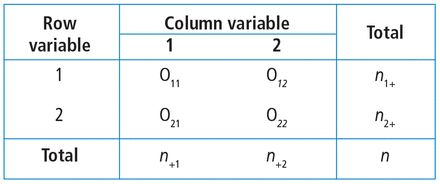
When there are 2 categorical variables, the data can be arranged in what is called a contingency table (Figure 1). Because both variables are binary (2 levels), it is called a 2 × 2 table. However, a contingency table can be generated for 2 categorical variables with any number of levels—in that case, it is called an r × c table, where r is the number of levels for the row variable and c is the number of levels for the column variable. The actual raw counts or frequencies are recorded inside the table cells. The cell counts are often referred to as observed counts and thus the notation (Oij) is used. The subscript i identifies the specific level of the row variable, and in this example it can equal 1 or 2 since the row variable is binary. Similarly, the subscript j identifies the specific level of the column variable and in this example it can equal 1 or 2 since the column variable is binary. Therefore, O11 represents the number of patients who have the row variable = level 1 and the column variable = level 1.
Example of a contingency table for 2 categorical variables, each with 2 levels (2 × 2 table).
In addition to the row and column variable cells, there are also the margin totals. These totals are either the row margin total (summing across the row) or the column margin total (summing down the column). For example, n1+ is the sum of the row where the row variable equal 1 (O11 + O12 = n1+). Finally, at the very bottom right corner is the grand total, which equals the sample size.
The goal is to test whether or not these 2 categorical variables are associated with each other. The null hypothesis (Ho) is that there is no association between these 2 categorical variables and the alternative hypotheses (Ha) is that there is an association between these 2 categorical variables.
The next step is to translate the generic form of the hypotheses into hypotheses that are specific to the research question. In this case, the null hypothesis is that mortality is not associated with lung morphology and the alternative hypothesis is that mortality is associated with lung morphology.
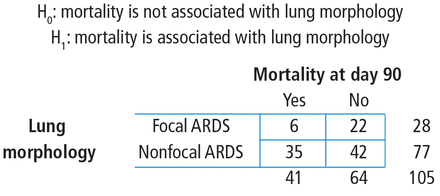
The contingency table cells can be populated with the numbers found in the article. It has our outcome of focus—mortality at day 90—both the count and the percent. The results are broken down by type of ARDS (focal vs nonfocal) as follows:
Focal ARDS = 6 patients (21.4%)
Nonfocal ARDS = 35 patients (45.5%).
From these numbers, we can build the contingency table that corresponds to the association among lung morphology (type of ARDS) and 90-day mortality (Figure 2).
Study-specific hypothesis, study frequency counts, and resulting 2 × 2 contingency table. Patient numbers are from the Mrozek study.1 ARDS = acute respiratory distress syndrome
First, the row variable is lung morphology, and it has two levels (focal vs nonfocal). Next, the column variable is 90-day mortality and it has 2 levels (yes vs no). Finally, the table must be populated, but be careful not to assume that there are no missing data. Begin with the cell counts: there were 6 focal ARDS patients and 35 nonfocal ARDS patients who died within 90 days. These two numbers populate the first column and result in a column total of 41. Next, use the reported percentages to calculate the row totals. Six is 21.4% of 28, so the first row total is 28. Thirty-five is 45.5% of 77, so the second row total is 77. If there are 28 patients with focal ARDS and 77 with nonfocal ARDS, then the grand total is 28 + 77 = 105. The remaining values can be obtained by subtraction. If there are 105 total patients and 41 die within 90 days, then 105 − 41 = 64 patients who do not die within 90 days and this is the second column total. Similarly, if there are 28 focal ARDS patients and 6 die within 90 days, then 28 − 6 = 22 patients who do not die within 90 days. Lastly, if there are 77 nonfocal ARDS patients and 35 die within 90 days, then 77 − 35 = 42 patients who do not die within 90 days. Now the contingency table is complete.
Once the contingency table is built, the question becomes, “Is lung morphology associated with 90-day mortality?” To answer that question, we need to know how many patients one would expect in each table cell if the null hypothesis of no association is true. When conducting a hypothesis test, one always assumes that the null hypothesis is true and then gathers data to see how well the data aligns with that assumption.
So one must calculate how many patients to expect in each of these cells if lung morphology is not associated with 90-day mortality. One way to address this question is to ask these 2 questions:
Overall, what proportion of patients die by day 90? Looking at the constructed contingency table, that answer would be 39%. This was calculated by taking the total number of patients who died by day 90 and dividing it by the total number of patients, 41/105 = 39%. This gives the overall proportion, based on the data, who would die by day 90.
How many of the focal ARDS patients would be expected to die by day 90? Now it is not overall, but rather we are limiting the question to the focal ARDS group. To obtain the answer, multiply the overall proportion of patients who die by day 90 by how many focal ARDS patients are in the study. Essentially, take the answer from the previous question and multiply it by the total number of focal ARDS, which is 28. The result is (41/105) × 28 = 10.9. Thus, if there is no association among long morphology and 90-day mortality, one would expect 10.9 focal ARDS patients to die by day 90.
Now 10.9 is a very specific answer for a specific contingency table, but the answer could be written in general terms. Basically, 3 numbers were used in calculating the solution: the row margin, the column margin, and the grand total. The general formula is the following:
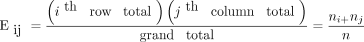
The notation Eij is used to represent the expected count assuming the null hypothesis of no association among the row and column variables is true. To calculate the expected count, take the ith row total times the jth column total and divide by the grand total.
In the lung morphology and mortality example, what is the expected number of deaths within 90 days among the nonfocal ARDS patients? This is the second row and the first column (E21). Applying the formula, one multiplies the total for the second row by the total for the first column and then divides by the grand total, (77 × 41)/105 = 30.1. This calculation is repeated for each of the 4 cells.
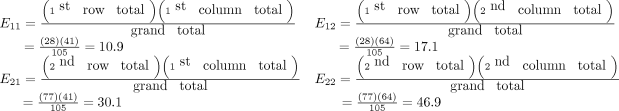
Because we now know the observed cell count and the expected cell count (under the null hypothesis), we can compare the observed and expected counts to see how well the data aligns with the null hypothesis. This is what the chi-square test does, and the test statistic is calculated as follows:
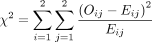
The sigma (Σ) means addition, so the calculation is performed on each individual cell in the contingency table and then the results are summed. A 2 × 2 table has 4 cells and thus 4 numbers will be summed. For each cell, the formula compares the observed to the expected. Basically, it computes how similar they are (that is the O minus E part). Because the differences will be positive for some cells and negative for others, the differences are squared to avoid cancellation when you add them. Finally, each squared difference is divided by the expected count to standardize the calculation.
Intuitively, if the observed counts (Oij) are similar to the expected counts under the null hypothesis (Eij), then these 2 numbers will be very close to each other. When taking the difference between them or subtracting them, the result is a small number. When squaring a small number, one obtains a really small number. And adding up a bunch of really small numbers results in a small number. So the test statistic is going to be small. That means that the resulting P value is going to be large. What is a P value? Think of it as an index of compatibility. How compatible is the data with the null hypothesis? Here, you get a large index of compatibility. That means that the data aligns nicely with the null hypothesis and one fails to reject the null.
Now, think about the alternative scenario. If the observed counts (Oij) are wildly different from the expected counts under the null hypothesis (Eij), then these 2 numbers will be quite different. When taking the difference between them or subtracting them, the result is a big number. When squaring a big number, one obtains a really big number, and adding up a bunch of really big numbers results in a large number. So the test statistic is going to be large. That means that the resulting P value is going to be small. And if you think of a P value as an index of compatibility, the data and the null hypothesis are not very compatible. That means that the data does not align nicely with the null hypothesis and one rejects the null. This is the general idea of the chi-square test. It assesses how compatible the data is with the null hypothesis that the 2 categorical variables are not associated.
To obtain the actual P value, the distribution of the test statistic (under the null hypothesis) is used to calculate the area under the curve for values equal to the test statistic or more extreme. The described test statistic has an approximate chi-square distribution with (r − 1)(c − 1) degree of freedom. Recall that r is the number of levels of the row variable and c is the number of levels of the column variable. Our example is a 2 × 2 table, so the test statistic has an approximate chi-square distribution with (2 − 1)(2 − 1) = 1 degree of freedom.
Now that the chi-square test has been fully described, the assumptions for the test must be discussed. It is important to know when you should or should not perform this test. The chi-square test assumes that observations are independent. This means that the outcome for one observation is not associated with the outcome of any other observation. This principle can be violated when multiple measurements are taken over time or when multiple measurements are taken from one patient.
Another assumption is that the chi-square large sample approximation just described is appropriate. In other words, no more than 20% of the expected counts (Eij) are less than 5. For a 2 × 2 table, how many cells do you have? Four. So if even one of those 4 happens to have an expected count less than 5, this assumption is violated. For a 2 × 2 table, none of the expected counts can be less than 5.
Returning to the lung morphology and mortality example, were the assumptions met? The data consist of 105 unique patients. Thus, we can assume that they are independent. The minimum expected count was 10.9, which is not less than 5. Therefore, the assumptions for the chi-square test are met. Next, the test statistic is calculated using the observed and expected counts. For each cell, subtract the expected count from the observed count, square it, and divide by the expected count. Then, add the 4 resulting numbers to obtain the test statistic of 4.92.
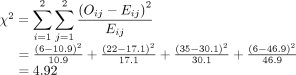
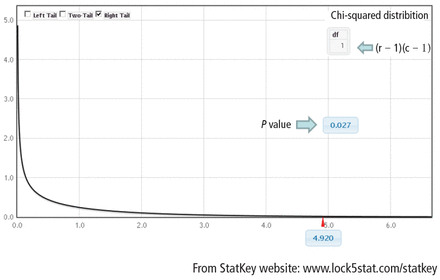
Finally, compute the area under the chi-square distribution with 1 degree of freedom, χ2(1), at the test statistic and values more extreme. In this case, values more extreme are values greater than the test statistic. Here, the area under the curve to the right of 4.92 is .027 (Figure 3). This is the P value, which indicates that the data and the null hypothesis have very low compatibility. In this example, the area under the curve to the right of 4.92 is .027 (Figure 3). This is the P value, which indicates that the data and the null hypothesis have very low compatibility. Thus, the decision is to reject the null hypothesis. The conclusion is that lung morphology is associated with 90-day mortality (P = .027). To describe that association, one looks at the contingency table and finds a reduction in 90-day mortality with focal patterns compared to nonfocal patterns (21.4% vs 45.5%, respectively). The P value reported in the article is .026. Our hand calculation was .027, which is slightly off due to rounding. In summary, the scenario is an investigation into the association among 2 categorical variables, and, thus, a test to consider is the chi-square test, if assumptions are met.
Chi-square distribution with 1 degree of freedom. Area under the curve at the test statistic of 4.92 and values more extreme equals the P value of .027.
In another example in the same study, the authors investigate whether any baseline characteristics are associated with lung morphology. For example, is neurology, specifically Parkinson disease (yes vs no), associated with lung morphology (focal vs nonfocal)? Again, the scenario is an investigation into the association between 2 categorical variables, so a chi-square test should be considered.
To start, build a contingency table arbitrarily placing lung morphology as the row variable and Parkinson disease as the column variable. Populate the contingency table based on the counts and percentages reported in the article (Figure 4). Next, check that the assumptions of the chi-square test are met. Are the observations independent? Again, because these are unique patients, we consider this assumption met. Since this is a 2 × 2 table, are all of the expected counts greater than 5? Calculations of the expected counts obtained the following: 1.1, 30.9, 2.9 and 84.1. Here, 2 of the 4 expected counts are less than 5. Therefore, methods that use large sample approximation, like the chi-squared test, may not be an appropriate choice.
Study-specific hypothesis and contingency table of lung morphology by Parkinson disease. Patient numbers are from the Mrozek study.1 ARDS = acute respiratory distress syndrome
Instead of using methodology that is an approximation, consider an exact test such as Fisher’s exact test. Again, refer to the contingency table where Fisher’s exact is going to calculate the exact probability (under the null hypothesis) of the observed data or results more extreme. This is the technical definition of a P value. It is, however, still quantifying how compatible the data are with the null hypothesis. The exact probability of a particular contingency table can be obtained using the hypergeometric distribution.
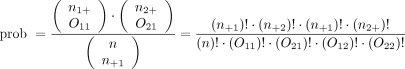
The symbols that resemble large parentheses are notations for a combinatorial. Because using combinatorials to calculate the probability is not user friendly, an equivalent version relies on factorials instead. Both techniques are presented above. Remember that the goal is to find the exact probability of the observed data or something more extreme.
The hypotheses are still testing whether these 2 categorical variables are associated with each other. In this particular example, we test if the proportion of patients with Parkinson disease is the same in the focal and nonfocal groups. Fisher’s exact test obtains its two-tailed P value by computing the probabilities associated with all possible tables that have the same row and column totals. Then, it identifies the alternative tables with a probability that is less than that of the observed table. Finally, it adds the probability of the observed table with the sum of the probabilities of each alternative table identified above, which results in the P value.
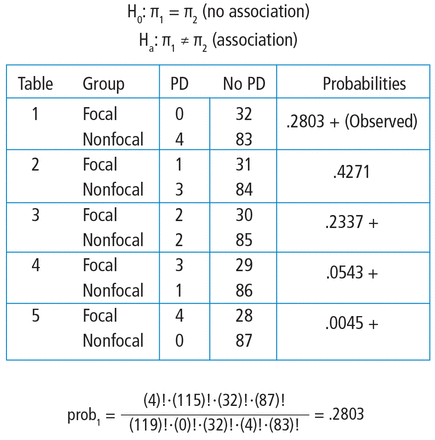
To explore each of those steps in detail, one must first enumerate how many tables can be built that all have the same row and column totals as the observed table. Figure 5 shows the 5 possible tables. Pick any one of the 5 2 × 2 tables; the margins are fixed. Each table has the same row totals, 32 focal and 87 non-focal, and each table has the same column totals: 4 Parkinson and 115 non-Parkinson. Then, for each table, calculate the probability of that table. Figure 5 shows this calculation for the first 2 × 2 table, which happens to be the observed table. The probability of the table observed in the study is .2803. Such a calculation is performed on each of the other tables.
Hand calculations of the Fisher’s exact test. Note that all tables have the same row and column totals. The probabilities of each table are calculated according to the hypergeometric distribution. Tables deemed “more extreme” (ie, with probabilities < the observed table) are indicated with a +. The P value is obtained by summing the probabilities of the observed table and those more extreme.
Next, one must identify the tables that have a probability smaller than the observed table. Here, we are looking for probabilities less than .2803. These are the tables deemed more extreme. Tables 3, 4, and 5 have probabilities less than .2803.
The final step is to sum the probability of the observed table and the more extreme tables (ie, those with probabilities < the observed table) (.2803 + .2337 + .0543 + .0045 = .5728). Thus, the resulting rounded P value is .57, which indicates a high level of compatibility between the data and the null hypothesis of no association. The decision is to fail to reject the null hypothesis and the conclusion is that the evidence does not support an association among lung morphology and Parkinson disease. In other words, there is insufficient evidence to claim that the proportion of Parkinson disease differs between the focal and nonfocal ARDS patients (0% vs 5%, P = .57). This matches the P value reported by Mrozek for this association.
The first objective of this article was to identify scenarios in which a chi-square or Fisher’s exact test should be considered. The general setting discussed was an investigation of the association between two categorical variables. Use of each test specifically depends on whether the assumptions have been met. Both of the examples used in our discussion happened to be binary, but that is not a restriction. Categorical variables can have more than 2 levels. All of the methods demonstrated for 2 × 2 tables can be generalized to r × c tables.
The second objective of this article was to recognize when test assumptions have been violated. For simplicity, most researchers adhere to the following: if ≤ 20% of expected cell counts are less than 5, then use the chi-square test; if > 20% of expected cell counts are less than 5, then use Fisher’s exact test. Both methods assume that the observations are independent. Could one use the exact test when the chi-square assumptions are met? Yes, but it is more computationally expensive as it uses all possible fixed margin tables and their probabilities. If the chi-square assumptions are met, then the sample size is typically larger and these calculations become numerous. Also, it does not have to be that large of a sample for the chi-square to be a good approximation and do it very quickly.
The final objective of this article was to test claims made regarding the association of 2 independent categorical variables. We included examples from the medical literature showing step-by-step calculations of both the large sample approximation (chi-square) and exact (Fisher’s) methodologies providing insight into how these tests are conducted as well as when they are appropriate.
Footnotes
This article is based on Dr. Nowacki’s presentation at the “Biostatistics and Epidemiology” lecture series created by Aanchal Kapoor, MD, Critical Care Medicine, Cleveland Clinic. Dr. Nowacki presented her lecture on January 10, 2017, at Cleveland Clinic.
Dr. Nowacki reported no financial interests or relationships that pose a potential conflict of interest with this article.
- Copyright © 2017 The Cleveland Clinic Foundation. All Rights Reserved.